Inputs : Integer n_D indicating the size of training data
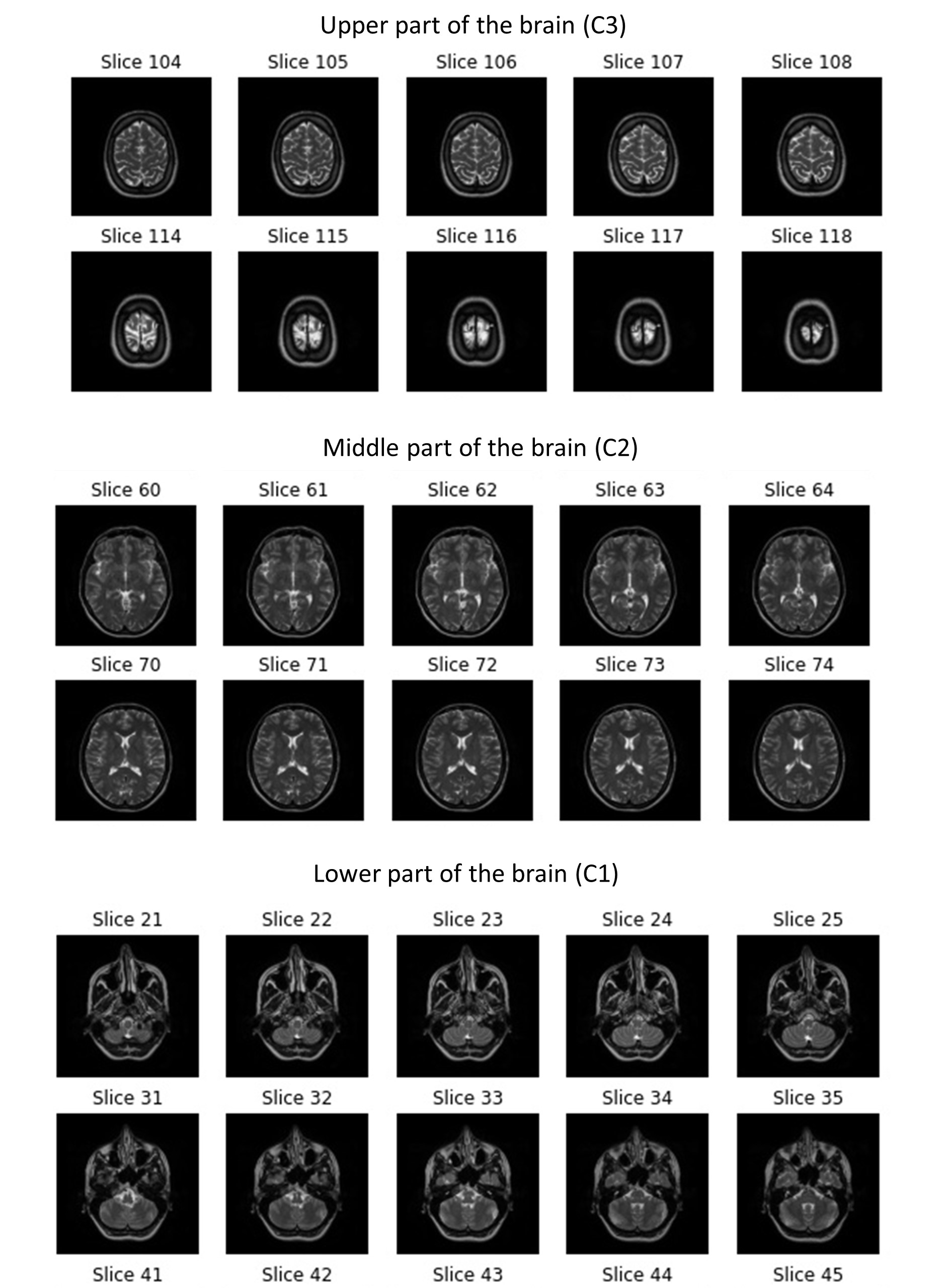
List C containing the different classes {“C1”, “C2”, “C3”}
String G representing the desired gender for the collection set: “M” for male or “F” for female
Integer n_L_A and n_L_B representing the lower and upper bound for subject range, respectively
Outputs : List D representing the sampled image slices for T2 and PD sequences
Step 1: Re-defining the lower bound L_B of subject range
if n_D < n_L_B then
n_L_B = n_D+n_L_A
endif
Step 2: Initializing necessary lists
Initialize S to {“T2”, “PD”}, representing the list of MRI sequences
Initialize D as empty two-dimensional list to store the sampled images from all sequences
Initialize R as empty one-dimensional list to store randomly sampled image slice numbers.
Step 3: Random sampling process based on stratified subject-wise splitting approach
for each sequence s in S, do
Open the directory of sequence s
Get and open the directory of gender G within sequence s directory
Initialize n_R to 0 for tracking the index in list R
for each class c in C, do
Initialize Y to the index of class c in C
Get and open the directory of class c to retrieve list P of subject directories
Initialize n_P to 0 for tracking the number of subjects processed
for each subject p in P within the specified range 〖n_L〗_A and 〖n_L〗_B, do
Step 3.1: Calculation of n_M for the count of sampled image slices
if index of s=0, which indicates s is “T2”, then
n_M = floor(n_D/n_L_B)+1
else
n_M = floor(n_D/n_L_B)
endif
Step 3.2: Calculation of n_T for the total number of image slices within subject p
Get and open the directory of subject p to fetch list I consisting of image files
Initialize n_T to 0
for each image file i in I, do
Increment n_T by 1
endfor
Step 3.3: Random selection of image slices from current subject p for sampling
Randomly generate n_M numbers within the range of n_T
Add the generated randomized numbers to list R
Step 4: Building the dataset of randomly sampled images
Open the directory of subject p to fetch list I consisting of image files
Initialize n_i to 0 for tracking the position of image file
for each image file i in I, do
Step 4.1: Selection of the correspondent image files for sampled slices
if n_i is inside the list R[n_R], then
Step 4.2: Feature extraction of selected image files
Read current image file i and store the result to X
Resize the shape of X to the desired image size of (224, 224, 3)
Step 4.3: Storing extracted features and the label of sampled images
Add X and Y of to D[index of sequence s in S]
endif
Increment n_i by 1
endfor
Increment n_R by 1
endfor
endfor
endfor
Output D, where D[0] contains the sampled data for T2 and D[1] for PD sequence